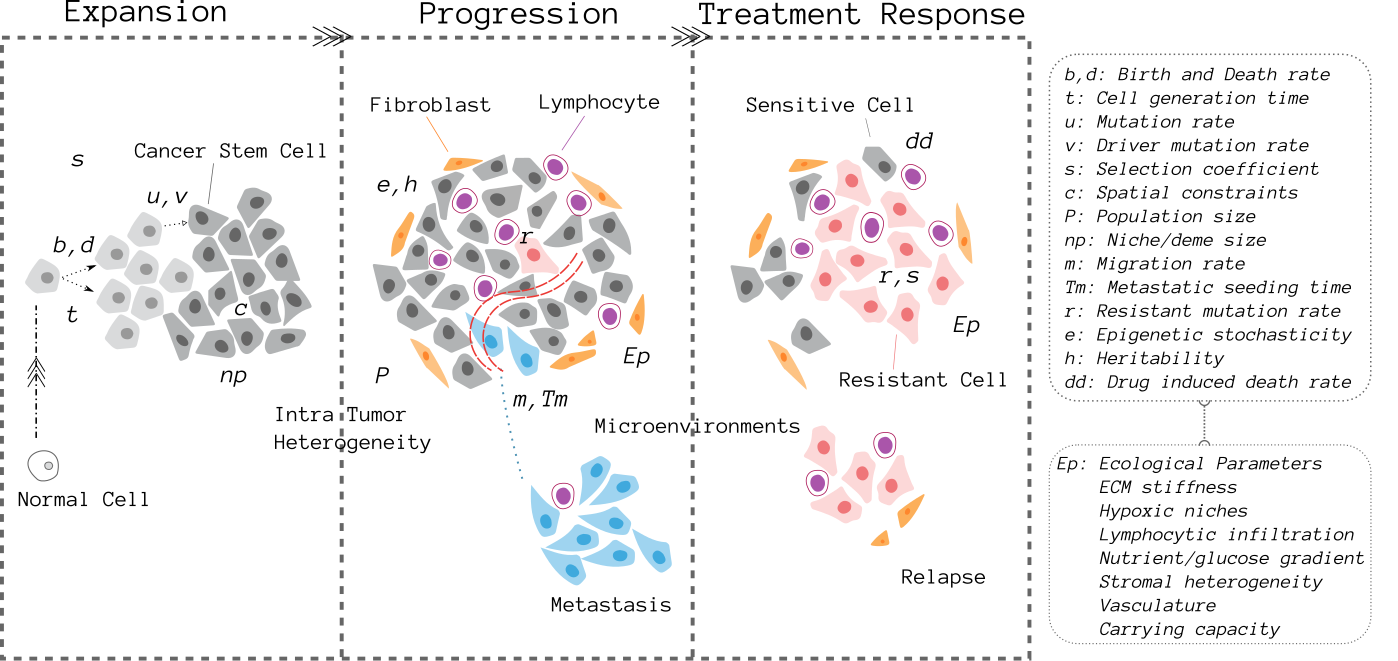
We innovate computational algorithms to quantify and model tumor heterogeneity (TH) from next generation sequencing (NGS) data, and leverage patterns of TH to statistically infer the kinetics of cancer.
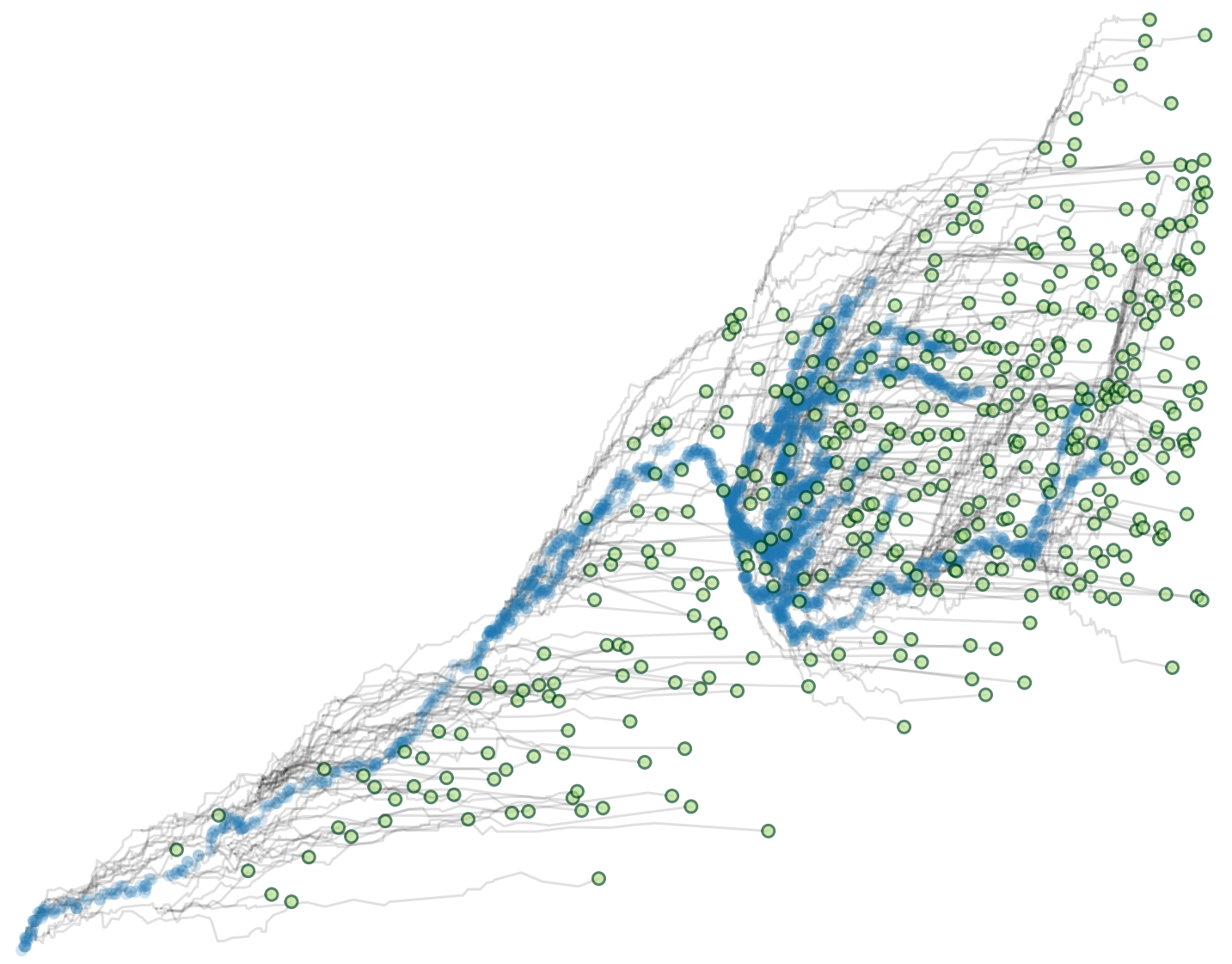
Tumor Clonal Dynamics: we perform data driven modeling of single-cell based spatial tumor growth, to reveal the principles that govern the relation between genomic heterogeneity, clonal dynamics, and sequencing detectability of somatic variants.
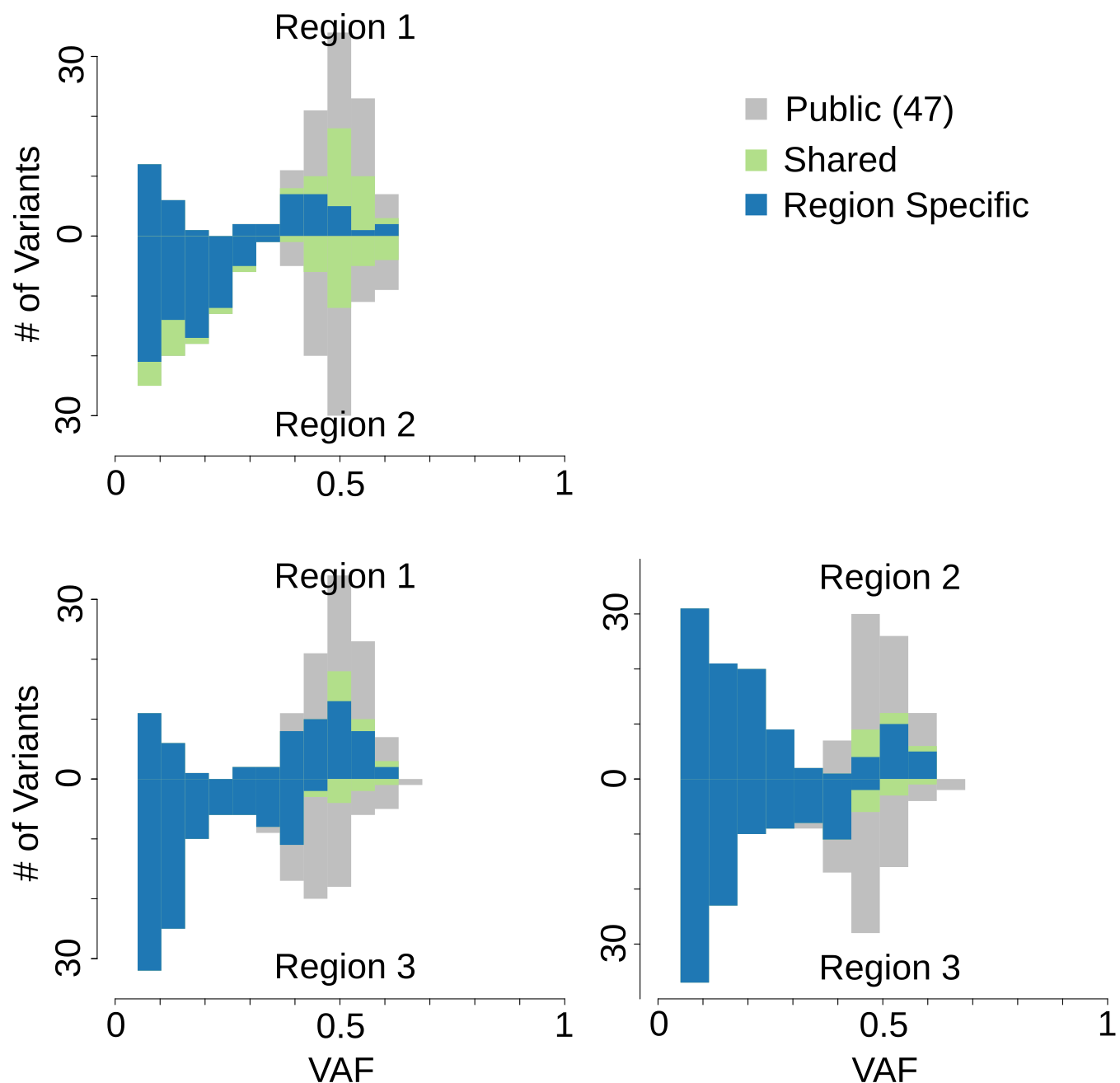
Computational Genomics in Tumor Heterogeneity: We push the limits in computational methods to achieve a high-fidelity quantification of (epi)genomic heterogeneity from tumor sequencing data (including CNA, SV, SNV, Gene Expression etc). Biases are inescapable but can be informative when we understand them better.
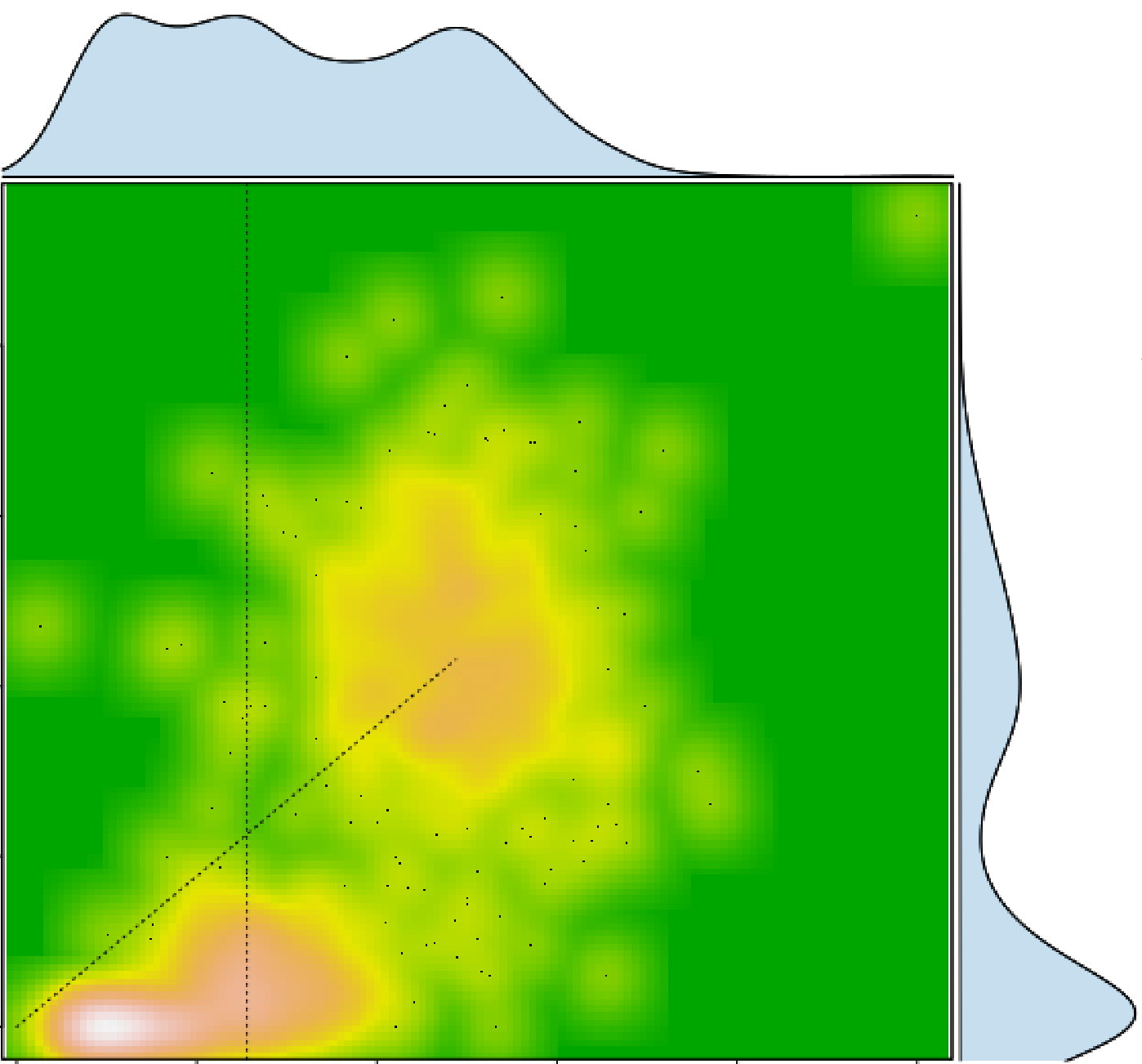
Visualizing Genomic Heterogeneity: Data visualizaion plays critical role in computational genomics. We develop visualization tools for tumor sequencing data to reveal the patterns of tumor heterogeneity, facilitate feature extraction, identify potential biases and inspire hypotheses regarding the dynamics of human cancers.
Our lab is affiliated with the Department of Laboratory Medicine and Pathology at the University of Minnesota, Twin Cities. We maintain vigorous cooperative research projects with pathologists, biologists, and mathematicians to bridge the scales among genomic variations, cellular behavior and tumor tissue patterns.
Open Post-Doctoral Associate Position:
The Sun lab is seeking a highly-motivated postdoc with a passion to innovate and discover in cancer data science. Our lab studies the dynamics of cancer by quantifying and modeling intratumor heterogeneity (ITH) from high-throughput sequencing data. We design computational algorithms to facilitate the translation of tumor heterogeneity into its evolvability. The potential research directions are listed below:
{Methods} for quantifying and visualizing intratumor heterogeneity
from NGS data;
{Computational and Mathematical modeling} of the spatial/temporal
tumor (epi)genetic heterogeneity, in consideration of tumor
microenvironments;
{Copy number evolution} in association with clonal dynamics.
The person who holds this position will receive hands-on mentoring of analytic and programming skills in advanced cancer genomics/bioinformatics and tailored training for career development. While the successful candidate is expected to get involved in the aforementioned research topics, his/her own research interests will be encouraged and promoted under the general scope of the lab. For more information on the position, applicants may directly email Ruping Sun (ruping at umn dot edu) with your CV.
Qualifications:
- A doctoral degree in bioinformatics, genetics, computer science, biostatistics, biomedical engineering, applied mathematics, biophysics, or other appropriate areas;
- Script language programming skills;
- Excellent written and verbal communication skills.
- Experience in cancer genetics, genomics and/or mathematical modeling (preferred).
- A record of publication and/or conference presentations.
Other Positions:
We welcome students and interns. Join us if
- you have good programming skills and want to step into the Cancer Genomics and Bioinformatics field to make an impact.
- you are trained in biostatistics, mathematics or biophysics and want to expand the potential of your work in the context of Cancer Genomics and Evolution.
- you are from biology background and like to learn more about bioinformatics in Cancer.
- you want to cross disciplines to increase your scientific skills.
Folks interested to work in the group directly email Ruping Sun (ruping at umn dot edu).